Diel protein-level regulation of plant cell processes
|
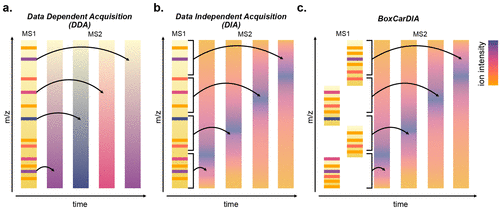
Using quantitative label-free proteomics, we aim to identify and characterize diel post-translational modification (PTM)-regulated plant cell processes. We recently developed a new data independent acquisition (DIA) workflow called BoxCar DIA that allows us to better quantify changes in the plant proteome.
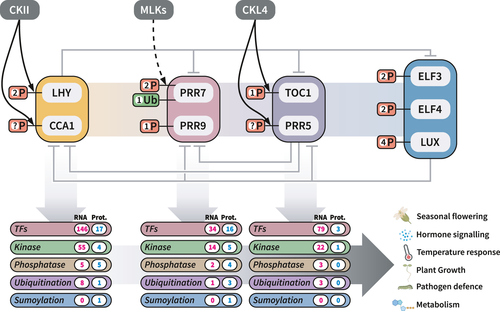
(Mehta, Scandola and Uhrig 2022 - Analytical Chem) Current status of the core plant circadian clock and associated outputs. The core plant circadian clock consists of a suite of transcription factors (LHY, CCA1, PRR7, PRR9, TOC1, PRR5, ELF4, ELF5, LUX) whose expression oscillates diurnally through the action of intertwined negative feedback loops. These core transcription factors are post-translationally modified, with some of the protein kinases identified. Core clock transcription factors have also been linked to changes in abundance of a large number of proteins belonging to various functional classes through transcriptomics and proteomics experiments. (Mehta, Krahmer and Uhrig 2021 - TPJ) |
Outstanding questions in diel plant cell regulation
Despite the circadian clock being extensively studied, there remains a number of gaps in our knowledge of how this occurs systemically across the regulatory landscape.
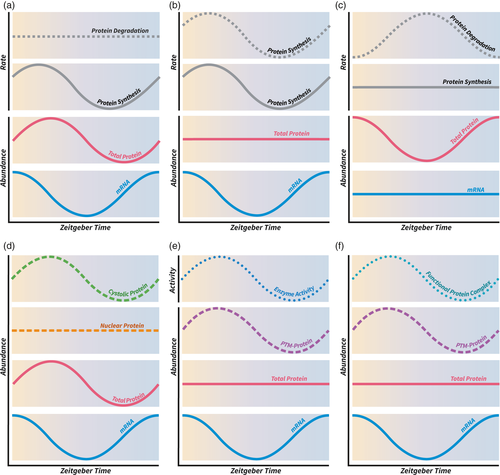
Estimates of protein and mRNA abundance obtained from simple proteomics and transcriptomics experiments have likely masked several underlying temporal dynamics that can only be revealed by studying protein turnover or by spatial proteomics, PTM-omics, or complexomics. (a) temporal oscillation of protein levels in phase with mRNA levels may result from oscillating rates of protein synthesis and constant degradation rates. (b - c) the apparent uncoupling of mRNA and protein-level oscillation may result from synchronized oscillating protein synthesis and degradation rates or stable rates of transcription and protein synthesis but oscillating degradation rates. Additionally, mRNA and protein-level data can mask dynamics in (d) subcellular localization, (e) enzyme activity and (f) protein complex formation, due to oscillating levels of post-translationally modified protein isoforms (PTM-Protein).
(Mehta, Krahmer and Uhrig 2021 - TPJ)
Estimates of protein and mRNA abundance obtained from simple proteomics and transcriptomics experiments have likely masked several underlying temporal dynamics that can only be revealed by studying protein turnover or by spatial proteomics, PTM-omics, or complexomics. (a) temporal oscillation of protein levels in phase with mRNA levels may result from oscillating rates of protein synthesis and constant degradation rates. (b - c) the apparent uncoupling of mRNA and protein-level oscillation may result from synchronized oscillating protein synthesis and degradation rates or stable rates of transcription and protein synthesis but oscillating degradation rates. Additionally, mRNA and protein-level data can mask dynamics in (d) subcellular localization, (e) enzyme activity and (f) protein complex formation, due to oscillating levels of post-translationally modified protein isoforms (PTM-Protein).
(Mehta, Krahmer and Uhrig 2021 - TPJ)
Targeted characterization of PTM modifying enzymes
Despite the identification of 1000s of PTM modified proteins, there remains a large gap in our understanding of which PTM modifying enzymes specifically catalyze which PTM events. Using a suite of functional genomic tools in combination with targeted proteomics, biochemistry and molecular biology our research efforts look to connect PTM modifying enzymes with specific substrates.